S26.5: New perspectives on the nature of species
Robert M. Zink1 & Jerrold I. Davis2
1J. F. Bell Museum, 100 Ecology Building, 1987 Upper Buford Circle, University of Minnesota, St. Paul, MN 55108 USA, fax 612 624 6777, e-mail rzink@biosci.umn.edu; 2L. H. Bailey Hortorium, Cornell University, Ithaca, NY14853, USA
Zink R. M. & Davis J.I. 1999. New perspectives on the nature of species. In: Adams, N.J. & Slotow, R.H. (eds) Proc. 22 Int. Ornithol. Congr., Durban: 1505-1518. Johannesburg: BirdLife South Africa.The biological species concept differs fundamentally from the phylogenetic species concept (PSC) and there is an active debate about which concept should be used in ornithology. We argue that the PSC should prevail. Taxonomic groups ranked as species differ under these two concepts because the BSC considers as conspecific those taxa that either can or are presumed able to interbreed, whereas the PSC considers groups with demonstrably separate histories to be species irrespective of their reproductive isolation status. Molecular systematic analyses now reveal cases in which patterns of history are incongruent with patterns of reproductive compatibility, because the ability to hybridise is an ancestral condition (plesiomorphy) that does not aid in estimating the historical course of diversification of the constituent taxa. Therefore, classifying taxa as species by their actual or presumed paths of potential gene exchange can be inconsistent with classifications based on recovered patterns of history. Such a classification is tolerated nowhere else in the taxonomic hierarchy. Therefore, phylogenetic species concepts are a decided improvement over the biological species concept because they accord logical primacy to patterns of historical diversification in the delimitation of species (we have a species concept empirically and theoretically applicable to allopatric populations). Phylogenetic species are the units of evolution required for hypothesis testing by evolutionists and population and conservation biologists alike. Adoption of the PSC means that systematics is a unified endeavour throughout the taxonomic hierarchy.
INTRODUCTION
Most biologists are aware of the long-standing controversy over species concepts. Nearly every field of biological endeavour, in fact, has contributed its views on how species should be delimited. Until recently, most arguments about species concepts were focused on how best to reconcile the dynamics of evolutionary change and the nature of interbreeding with the Linnaean system of binomial nomenclature. Put more simply, the debates centered on the question of whether it was better to classify nature by being a ‘splitter’ or a ‘lumper.’ Our goal here is to show that the debate over species concepts has been materially changed in recent years by contributions from two fields. And, that these new developments together lay the framework for a species concept, the phylogenetic species concept (PSC), that has widespread generality and that, in our opinion, is a decisively better choice than the ‘biological species concept’ that prevails in ornithology (AOU 1998).
The methods and goals of the school of systematics termed phylogenetic systematics (Hennig 1966), or ‘cladistics,’ have become widely accepted and they provide an objective approach to the reconstruction of evolutionary histories of species and higher-level groupings, and a sound set of principles for generating classifications. It has become clear, however, that the cladistic approach is applicable ‘below’ the traditional species level, which has broad implications for the species debate. Thus application of phylogenetic principles at and below the biological species level is the first important recent addition to the species debate. The second contribution comes from molecular systematics. Analyses of DNA polymorphisms within and among populations 1) reveal or confirm groups of individuals within traditional biological species that have had separate evolutionary histories, and 2) allow reconstruction of historical (phylogenetic) relationships among those groups. These two recent developments have changed the species debate by calling into question how, or whether, cladistic rules of classification should be applied to groups within traditional species whose existence and phylogenetic relationships are discoverable by molecular methods.
The so-called biological species concept (BSC), developed principally by Dobzhansky, Mayr, and associates during the first third of this century (Dobzhansky 1937; Mayr 1942, 1963), has arguably prevailed for the past 50 years, and has become firmly entrenched in most textbooks: ‘Species are groups of actually or potentially interbreeding natural populations, which are reproductively isolated from other such groups’ (Mayr 1942:120). Nonetheless, challenges to the BSC have been made and are occurring in many places (Ehrlich 1961; Cracraft 1983; Donoghue 1985; McKitrick & Zink 1988; Frost & Hillis 1990; Whittemore 1993; Luckow 1995; Mayden & Wood 1995; Zink 1997a; we do not review criticisms of the BSC here but refer readers to these citations). In ornithology for example, a traditional bastion of support for the BSC, the leading textbook (Gill 1995) gives consideration to alternative species concepts. This was not present in the previous generation of textbooks. Whereas some formal and established classifications of birds adhere to some version of the BSC (e.g., AOU 1998), others, such as the Dutch committee for avian systematics, are ‘reluctant to use the Biological Species Concept as the framework for the delimitation of bird species and has adopted a more cautious phylogenetic approach to delimit species’ (Sangster et al. 1997:21). Thus the BSC is being actively debated in ornithology, which is also true for other groups of organisms (Davis 1995).
Phylogenetics and Species Concepts
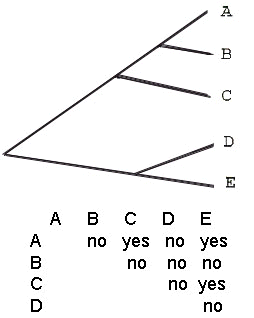
Phylogenetic systematics (Hennig 1966) requires that classifications be faithful to recovered patterns of evolutionary history. That is, the classification should reflect the reconstructed genealogy, which is estimated by the pattern of shared-derived characters. The phylogenetic method is familiar to most biologists as it is applied ‘above’ the species level. However, the goal of representing historical patterns at and ‘below’ the species level in classifications represents a major area of conflict between the PSC and the BSC. For example, assume that one had recognised five groups of individuals by some suite of characters, and had reconstructed their phylogenetic history (Fig. 1). Furthermore, assume that their ability to hybridise and produce fertile young in a hybrid zone was either known, or inferred by experts familiar with the group. The tree shows that the pattern of reproductive compatibility is incongruent with the known pattern of evolutionary history. That is, grouping taxa by the possession of the primitive ability to hybridise results in different groupings than implied by the tree. The evolution of reproductive isolation is just one component of diversification, and it may not occur concomitantly with other aspects of differentiation (Rosen 1979; Bremer & Wanntorp 1979). As Bush (1982:119) commented, ‘reproductive isolation is after all the end product, not the cause, of speciation.’ Under the BSC, one would consider as the same species all individuals in groups A, C, and E. A, C, and E differ in other character suites, which allow the phylogenetic pattern depicted in Fig. 1 to be resolved, and they should not be pooled according to the principles of phylogenetic systematics because groupings based on retention of an ancestral characteristic (ability to interbreed) yields species classifications that misrepresent the cladistic pattern (taxon ACE is paraphyletic). Put another way, to place (potentially) hybridising taxa in a common species is analogous to arguing that two stars should be recognised as only one star today because there is a chance of their colliding within the next 500 million years; that potential is interesting, and worth considering, but we recognise two stars today, each with a unique history and a distinct name. Many avian taxa with distinct evolutionary histories are pooled as conspecific under the BSC because they might fuse in the future as a consequence of actual or potential hybridisation (Zink & McKitrick 1995).
The connection between the species debate, on the one hand, and phylogenetics and molecular systematics above the species level, on the other, is straightforward. Traditionally avian taxonomists routinely described taxa within species, such as subspecies, but they were not overly concerned with the pattern of evolutionary relationships among these subspecific taxa. For example, in ornithology there were many monographic studies of biological species, such as the North American flickers (Short 1965) and juncos (Miller 1941) but these authors primarily were concerned with delimiting taxa and assessing whether they could interbreed, not how they were related hierarchically. In fact many characters that diagnose taxa at this level appear low in phylogenetic information content. Therefore, it was not recognised that application of the BSC could lead to situations in which taxa grouped according to their actual or potential reproductive compatibility might conflict with those grouped according to their historical patterns of evolutionary relationships (Fig. 1). Molecular analyses provide characters that not only confirm historical groups but more importantly facilitate reconstruction of phylogenetic pattern. The Fox Sparrow Passerella iliaca provides an example; 18 subspecies were recognised among four basic plumage groups but there was relatively little attention paid to how the groups might be related (Swarth 1920), owing to the lack of a phylogenetic method at this taxonomic level. MtDNA studies (Zink 1994) confirmed the existence of four groups and hence four phylogenetic species, but more importantly showed that the recovered phylogenetic pattern (Fig. 2) conflicts with the pattern of hybridisation because a prominent hybrid zone exists between two non-sister taxa (schistacea and megarhyncha). Thus putting the two hybridising fox sparrow groups into a species to the exclusion of the others creates a non-historical grouping (megarhyncha), one of no predictive value and one inappropriate for biogeography, phylogeny, comparative studies and the enumeration of biodiversity (Cracraft 1997).
Epistemologically, modern systematic biology places primary emphasis on objectively recovered evolutionary patterns, and not processes associated with these patterns (Graybeal 1995), as the basis for classification. For example, a steady stream of papers is being produced in which the phylogenetic framework is used to deduce evolutionary processes. To adopt the BSC is to reverse these endeavours at the species level (such as emphasising the hybridisation between schistacea and megarhyncha), which means that some investigators have different sets of rules for classifying taxa below and above the ‘species level’. That is, phylogenetic patterns are used above the species level to create classifications and information on reproductive compatibility is added below the species level (and the concept of synapomorphy abandoned). There is no need for two sets of rules to classify taxa. For example, under the BSC one should consider schistacea and megarhyncha as the same species because they have not diverged to the point at which they are reproductively isolated, despite the fact that they have mtDNA and morphological differences (although the AOU [1998] recently recognised that stable hybrid zones suggest separate species status for hybridising taxa, determining the stability of a hybrid zone can take thousands of generations and is therefore a dubious criterion). Hence the BSC ‘weights’ the process of mate choice over the pattern of divergence. The simple phylogenetic explanation is that reproductive compatibility has been retained in two non-sister phylogenetic species, and not that hybridisation is indicative of conspecific status, the latter of which should imply historical coherency. Just as we would not unite birds and mammals simply because they are ‘warm blooded’ we would not require our classification of schistacea and megarhyncha to violate rules of phylogenetic classification.
Although there are many competing species concepts (Paterson 1985; Templeton 1989; Mayden & Wood 1995; Zink 1997a), we focus on a new class of species concepts (Rosen 1979; Nelson & Platnick 1981; Cracraft 1983; Nixon & Wheeler 1990; Davis & Nixon 1992) that have been crafted with the express goal of providing faithful representations of historical patterns of diversification. In contrast to the BSC, reproductive compatibility is not considered evidence of conspecificity - species can and do hybridise without losing species status (of course extensive hybridisation will result in an inability to recognise once-distinct species, although this requires long periods of time [Zink & McKitrick 1995]). Furthermore, these new species concepts apply to allopatric populations by using directly observable character evidence to delimit species - not by filtering character differences and hypothesised mate choice behaviours through individual taxonomists' experience.
Two principal approaches to species concepts rooted in phylogenetics have emerged (Nixon & Wheeler 1990). The first (e.g., Eldredge & Cracraft 1980; Nelson & Platnick 1981; Cracraft 1983, 1989; Nixon & Wheeler 1990, 1992; Davis & Nixon 1992) is based on divergence of gene pools (the phylogenetic species concept, or PSC), whereas the second (e.g., Mishler & Donoghue 1982; Donoghue 1985; Mishler 1985; Mishler & Brandon 1987; de Queiroz & Donoghue 1988; Baum 1992) is a set of monophyly-based species concepts. A third category, based on patterns of gene coalescence rather than on descent relationships among the constituent organisms of a species, has been proposed (Avise & Ball 1990; Baum & Shaw 1995; Mallett 1995; Avise & Wollenberg 1997), but we do not discuss it further here (see Davis 1996, 1997).
Genetic divergence following the cessation of gene exchange among populations or complex population systems will eventually result in the development of diagnostic differences, such as unique morphological characters or mutually exclusive sets of alleles or DNA sequences at one or more loci. Population systems that can be discriminated from others by such character differences are recognised as phylogenetic species (PS). The term ‘phylogenetic’ is applied in this case because these groupings, having ceased to exchange genes with other such groups, are the minimal groupings among which there is evidence of phylogenetic structure. It does not imply that phylogenetic methods are used for their discovery (Davis & Nixon 1992; see below). In contrast, the absence of diagnostic character differences among populations within a PS is suggestive of continuing gene exchange, however infrequent, and relationships among such populations therefore appear to be reticulate (the status quo within populations), rather than hierarchic. To put the point differently, if no diagnostic differences are discovered among several local (allopatric) populations it is not clear that they are independently evolving taxa (gene flow might still exist), and they may be regarded provisionally as elements of a single inclusive population system. Hence, phylogenetic species lie at the boundary between reticulate and hierarchic descent relationships, and diagnostic character combinations represent the means by which such groups can be identified. Because the isolating barriers of biological species are themselves character differences, the PSC recognises the boundaries between biological species, as well as a larger set of boundaries; in short, a PS may be either coextensive with or less inclusive than a biological species, if the available evidence is interpreted equally strictly.
The various monophyly-based species concepts share the goal of delimiting species whose members either share a more recent common ancestor with each other than with nonmembers, or who are more closely related to each other than they are to organisms in other species. For example, consider three local populations, A, B, and C, between which there are occasional episodes of gene flow. At a later time, geographic barriers to gene flow develop and the three populations cease to exchange genes with each other. If a character evolves within population C that does not confer reproductive isolation, it will be recognised as a phylogenetic species, but will still be regarded as belonging to the same biological species as populations A and B. If the character that evolves within population C constitutes the basis of a reproductive isolating barrier, it will be also be recognised as a biological species. In both of these cases, populations A and B will be regarded as belonging to the same phylogenetic species as well as to the same biological species. Most monophyly-based species concepts will agree with the PSC in recognising population C as a distinct species in both of these cases, for proponents of the various monophyletic species concepts interpret its unique character as evidence of monophyly (Donoghue 1985; de Queiroz & Donoghue 1988). However, neither population A or B has a uniquely derived character, nor does the larger grouping that includes both populations, so the individuals within these populations are regarded as belonging to no species, and these two populations would be recognised as a ‘metaspecies’ (Donoghue 1985; de Queiroz & Donoghue 1988).
Implications of Phylogenetic and Biological Species
In part proponents of the BSC and PSC differ over the level at which species names should be employed. However, a marked departure between the two concepts occurs when the pattern of reproductive compatibility conflicts with the pattern of evolutionary history (as in the example discussed above of the Fox Sparrow). Whereas the BSC emphasises the potential for future gene flow, the PSC emphasises the actuality of differentiation in form and other characters - evolutionary history. Thus the debate over species concepts is not a semantic debate about whether to be a splitter or lumper, rather, it concerns when and how to apply phylogenetic principles of classification. Below we explore other topics that have been raised concerning the conflict between the BSC and PSC.
Defences of the BSC and Criticisms of the PSC
Although many papers have criticised the BSC and its use in ornithology, there are been relatively few ‘defences’ of it.The recent AOU Check-list of North American Birds (AOU 1998) responds to criticisms of the BSC, as well as offers some criticisms of the PSC.
Many have pointed out the fact that ranking allopatric populations is a flaw with the BSC, as it requires subjective guesses as to whether there are mate choice mechanisms in place that would result in assortative mating if sympatry were achieved. The AOU Check-list Committee (1998) states that ‘Quantified study of vocalisations and detailed investigation of genetic distances..’ can resolve the species status of allopatric populations. Certainly this information is relevant but hardly definitive in what should be a phylogenetic exercise. Surprisingly this is recognised by the AOU, which states (xiv-xv) ‘Moreover, we regard as indefensible the identification of species by what are essentially phenetic criteria.’Genetic distances and multivariate analysis of vocalizations are utterly phenetic. Hence the problem of allopatric populations looms as large as ever.
The treatment of hybrid zones under the BSC has been inconsistent (Zink and McKitrick 1995).The AOU Check-list Committee (1998) now recognises that Mayr (1982) noted that a stable hybrid zone implies that the two hybridising forms are separate species, as it signifies a lack of free interbreeding. Yet, given this philosophy, this same committee (1998:542) considers the Myrtle Warbler Dendroica coronata and the Audubon's Warbler D. auduboni the same species because ‘free interbreeding occurs in a narrow hybrid zone ...’.Narrow hybrid zones are likely stable, revealing again the inconsistent treatment of hybrid zones by advocates of the BSC.
The AOU Check-list Committee (1998) is concerned that the PSC is not biological, apparently considering irrelevant the remarks of Zink and McKitrick (1995) to the contrary. In fact the Check-list committee (1998) remarks ‘This is revealed clearly by the fact that the PSC can apply equally well to either animate or inanimate objects...’. However, in preparation of the faunal list of North American birds, this same committee reviewed and included results from literature that has involved ‘application of phylogenetic or cladistic approaches’. The contradiction here is striking as it has been widely appreciated that cladistics applies to inanimate objects at all levels (Nelson and Platnick 1981). Hence this criticism is meaningless and irrelevant. Phylogenetic species are every bit as 'biological' as biological species, and the AOU's (1998) statement to the contrary reveals a fundamental lack of understanding of entities analysed by phylogenetic methods.
Methods for Discovery of Phylogenetic Species
Davis & Nixon (1992) describe a procedure, population aggregation analysis (PAA), by which individuals within and among localities are sorted into species by directly examining character evidence, and without reference to the potential for gene flow between groups. They further point out that phylogenetic methods per se are not necessarily a part of the description of phylogenetic species via PAA, and in that sense the ‘phylogenetic’ in phylogenetic species concepts is potentially misleading; phylogenetic species are the taxa that once recognised may be used in phylogenetic analysis. They are not further divisible by multi-character analysis. Unfortunately, PAA has not been rigorously applied to any group of birds; it is illustrated with a hypothetical example in Zink and McKitrick (1995).
Several misconceptions about phylogenetic species exist in the literature. For instance, does acceptance of the PSC mean that separate sexes, or separate instars of conventionally recognised insect species, or even individual organisms are separate species because these ‘groups’ are diagnosable? Avise & Ball (1990) suggested that the PSC leads to the inevitable conclusion that individual organisms could be separate phylogenetic species because analyses of DNA likely would show that every individual is ‘diagnosable.’ This misrepresents the goal of the PSC, which is to discover fully differentiated population systems (here we see parallels between phylogenetic and ‘evolutionary species’ [Mayden & Wood 1995]), not to delimit arbitrarily identifiable groups within populations. Phylogenetic species typically will be based on analyses of multiple characters, which lead to hypotheses about species limits. In such analyses there will often be character conflicts, which PAA and cladistic methods accommodate. Species limits are hypotheses, and phylogenetic species are tested directly by analyses of additional characters. Moreover, the central role of the population in the PSC clearly mitigates against the acceptance as distinct species of individual organisms or different developmental stages or separate morphs that coexist within populations.
An avian example involves the Fox Sparrow Passerella iliaca discussed above. One could ask why more than four phylogenetic species were not recognised, because for example within one of the four groups, iliaca, 20 distinguishable haplotypes were found among 72 individuals (Fig. 3). Why were not individuals possessing a distinct haplotype each considered a separate phylogenetic species by virtue of being genetically diagnosable? The answer is that there was no other evidence in the form of scorable characters that supported finer or less inclusive groups of individuals as having had a distinct evolutionary history. The four mtDNA lineages correspond to groupings recognised on morphological grounds as separate species and, hence, the evidence converges on a hypothesis of four phylogenetic species. Variation alone does not prevent discovering historical taxa, nor does it mean that one biological species will automatically be multiple phylogenetic species. For example, one might use highly variable microsatellite markers and find many ‘overlapping’ groups of individuals within a population sample. However, using PAA, the population when compared against other population samples would be scored as polymorphic, and the variation at microsatellites would not be constitute evidence for phylogenetic species (Davis and Nixon 1992). Groups of individuals that have had separate histories will likely have several character systems that all capture (and agree on) this unique history.
Does a PSC Imply Greater Ease in Grouping Individuals into Species Taxa?
Mayr (1982) noted that one can identify two procedural steps in delimiting species - grouping individuals into taxa, and ranking these taxa as some level in the taxonomic category. It is often difficult to group individuals into clearly distinct taxa, owing to sexual dimorphism, age-related differences, and polymorphisms, irrespective of what species concept is followed. For example, many avian taxonomists have struggled to define subspecies, invoking rules of 75% or 95% conformance; this shows the difficulty of placing individuals into taxa because of the variation inherent in natural populations. Similarly some phylogenetic species are differentiated only slightly more or less sharply than some threshold level that is adopted (Cracraft 1983). This difficulty does not invalidate grouping individuals into taxa, however, for either species concept. Thus the shift in paradigm between the PSC and the BSC does not involve grouping individuals into taxa, rather it involves ranking taxa at the species level. The PSC calls for species limits to be consistent with the smallest groups of individuals identifiable as having had independent histories, whereas the BSC superimposes information about actual or potential interbreeding onto group histories and can therefore include multiple phylogenetic species in a single biological species (allowing paraphyletic taxa; Fig. 2).
The Role of Populations and Gene Flow in Species Concepts
The meaning of population is related to the notion of lineage. If two apparent ‘populations’ exhibit no character differences, then in effect, a null hypothesis is that they have not diverged. The null hypothesis is rejected if a heritable character difference(s) is found. This notion of extended genealogical systems (i.e., groups united by gene flow in nature) should be of great interest to evolutionists and population geneticists, for it is reflective of the historical limits of gene flow. The BSC, in contrast, can group populations between which there is evidence that gene flow as ceased, and thus recognises composite taxa - biological species - that are united only by the potential for gene flow to occur.
The neo-Darwinian synthesis clarified the role of gene flow within and among populations as a key component of the evolutionary process. Biological species, by definition, were groups that could not exchange genes with each other, and thus it seemed natural to regard each of these groups as the arena within which evolution proceeded. Dobzhansky (1937) emphasised the importance of barriers to gene flow, and species came to be seen as the most extensive communities of individuals that could exchange genes. This criterion of species membership will delimit the largest possible community within which gene exchange might occur, but it will often include separate population systems that have had independent histories for considerable amounts of time and that have themselves yielded lineages and sublineages that have differentiated from each other in the absence of interbreeding among them; all that unites the disparate elements of these biological species is the possibility that they may exchange genes at some future time. Systematists and other evolutionary biologists have recognised that a crucial level of organisation exists above the local deme and below the biological species, that being individual populations and groups of populations that occasionally exchange genes with each other, but have ceased to exchange genes with other such extended population systems, as evidenced by the evolution of unique character combinations (but not necessarily in mate choice or other attributes of reproductive biology). Gene flow occurring between local populations of a phylogenetic species can be rare or frequent, but the existence of diagnostic character combinations is prima facie evidence that gene exchange among phylogenetic species does not occur (Davis & Nixon 1992). Thus, the goal of the BSC is to identify the most inclusive population systems among which there is no potential for gene flow to occur, whereas the goal of the PSC is to identify the most inclusive population systems among which gene flow does not occur.
Many biologists are interested in the potential pathways that genes might take among taxa. The BSC yields units that potentially capture such pathways. However, the paths that genes might or do take is best studied first from the correct representation of historical relationships. That is, mapping the potential route of genes onto the correct historical hypothesis provides a framework for studying the evolution of reproductive isolation. For example, Cracraft (1989) noted that a hybrid zone and resultant introgression could be attributed to ‘secondary contact’ if the taxa involved were not sister taxa. Thus, rather than obscuring the study of gene flow, adoption of the PSC enriches its analysis.
Numbers of Bird Species and Molecular Systematics
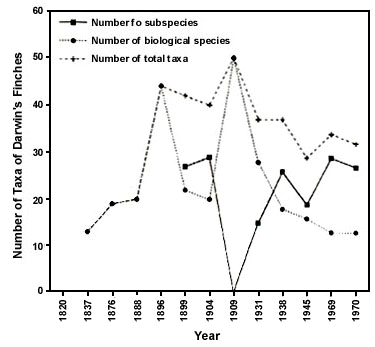
One of the stated virtues of the BSC was that it reduced the number of recognised species, subsuming many allopatric populations, often considered geographic or ecological replacements, into more inclusive biological species (Mayr 1942). Thus the number of recognised bird species dropped from about 20,000 to 8,500. That is, many traditionally recognised species were demoted to subspecies because investigators deemed that they would hybridise with near relatives, if ever they were to become sympatric. The classification of Darwin's finches reveals the impact of the transition to the polytypic BSC (Fig. 4). Under a PSC, many of the taxa formerly considered subspecies would be recognised as independently evolving, basal evolutionary taxa or phylogenetic species. Fig. 4 suggests that the impact of a PSC would result in an upper bound less than the number of total taxa, as likely many subspecies would be found to be arbitrary divisions of clinal patterns of variation and not recognised as phylogenetic species. It is not likely that the PSC would more than double the number of avian species (Mayr 1993), but neither can there be an a priori limit to the number of species. We suggest that use of the PSC for birds would result in a classification that would provide a more objective comparison of bird species diversity with other, far more speciose groups such as plants and insects that are essentially classified with a phylogenetic concept.
Martin (1996) suggests that molecular methods will reveal many phylogenetic species hidden from taxonomists by lack of morphological change. Indeed, if true, field studies could be hindered if species could not be recognised by external phenotypic criteria. However, few existing molecular studies of birds suggest phylogenetic divisions within biological species for which there is no concomitant morphological evidence (Fig. 2). Genetic studies of Darwin's finches (Yang & Patton 1981) suggest that one of the commonest and most widespread species, Certhidea olivacea, consists of two historical taxa, but not 10 or 100. These two groups belong to different subspecies, indicating that differentiation already had been recognised and formally named via the taxonomic code. Only 1 of over 25 molecular studies of continental North American birds revealed a significant historical break that was not also apparent in morphological attributes (Zink 1997b). Conversely, many morphologically-inferred subspecies of birds are likely not historically significant lineages (Ball & Avise 1992). For example, in the most highly polytypic species in North America, the Song Sparrow Melospiza melodia, mtDNA analyses (Zink & Dittmann 1993; Fry & Zink 1998) do not reveal discrete historical groupings, and the extensive morphological variation appears clinal (Aldrich 1984). Hence, none of the 34 subspecies of the Song Sparrow appear to qualify as separate phylogenetic species (under the PSC there would not be a subspecies category). Thus, molecular studies will not greatly increase the number of recognised avian species (Zink 1996); mostly they will reveal which already named subspecies are actually phylogenetic species.
The Biological Species Concept: A retrospective salute
Application of phylogenetic principles consistently throughout the taxonomic hierarchy amounts to abandonment of the BSC. Rather than detrimental we view this as advantageous. With systematics of most groups largely transformed to a phylogenetic context, it is time to turn to the species level itself. Proper orientation of taxa into species will in fact facilitate study of the very aspects that the BSC considers its cornerstone. For instance, the question is why have schistacea and megarhyncha retained the primitive ability to hybridise relative to their true sister taxa (Fig. 2)? A correct phylogenetic classification sets the framework for decisive experiments.
The future of the species debate is uncertain. Some schools of thought maintain preeminent importance for the BSC whereas others hold it to be eroded to the state of uselessness. Thus, even the debate over the debate is controversial, but in retrospect, the BSC has focused attention on variation within populations, genetic models of adaptation, the significance of hybridisation in speciation, and the process of mate choice. However challenges from molecular and phylogenetic systematics have seriously eroded the BSC; even conservation biologists define evolutionary significant units that are phylogenetic entities (Moritz 1994). Ultimately this will complete the transition from evolutionary to phylogenetic thinking at ‘lower’ taxonomic scales, by making species equivalent to the entities we have described here as phylogenetic species. Until then the revolution in systematics and classification started by Hennig remains incomplete.
ACKNOWLEDGEMENTS
We thank many individuals for discussing these issues with us over the past 10 years (without implying agreement on issues). Both authors acknowledge funding support from the US NSF. R. Shaw, S. Weller, A. Kessen, and S. Naeem provided useful comments on versions of the manuscript. We thank G. Barrowclough for assistance in preparing Fig. 4.
REFERENCES
Aldrich, J.W. 1984. Ecogeographic variation in size and proportions of Song Sparrows (Melospiza melodia).Ornithological Monograph No. 35, Washington, D. C. American Ornithologists' Union.
American Ornithologists' Union. 1998. Checklist of North American birds, 7th ed. American Ornithologists' Union, Washington, DC.
Avise, J.C. & Ball, R.M. 1990. Principles of genealogical concordance in species concepts and biological taxonomy. Oxford Surveys in Evolutionary Biology 7: 45-67.
Avise, J.C. & Wollenberg, K.1997.Phylogenetics and the origin of species.Proceedings National Academy of Sciences USA 94: 7748-7755.
Ball, R.M. Jr & Avise, J.C. 1992. Mitochondrial DNA phylogeographic differentiation among avian populations and the evolutionary significance of subspecies. Auk 109: 626-636.
Baum, D.A. & Shaw, K. L. 1995. Genealogical perspectives on the species problem. In: Hoch, P. C. & Stepenson, A. G. (eds) Experimental and Molecular Approaches to Plant Biosystematics. Monographs in Systematic Botany, Missouri Botanical Gardens 53: 289-303.
Bremer, K. & Wanntorp, H.-E. 1979. Geographic populations or biological species in phylogeny reconstruction? Systematic Zoology 28: 220-224.
Bush, G.L. 1982. What do we really know about speciation?In: Milkman, R. (ed) Perspectives on Evolution. Sunderland; Sinauer Associates: 119-128.
Cracraft, J. 1983. Species concepts and speciation analysis. Current Ornithology 1: 159-187.
Cracraft, J. 1989. Speciation and its ontology: The empirical consequences of alternative species concepts for understanding patterns and processes of differentiation. In: Otte, D. & Endler, J.A. (eds) Speciation and its consequences. Sunderland; Sinauer Associates: 28-59.
Cracraft, J. 1997. Species concepts in systematics and conservation biology: an ornithological viewpoint. In: Claridge, M.F., Dawah, H.A. & Wilson, M. R. (eds) Species the units of biodiversity. London; Chapman and Hall. in press.
Davis, J.I. 1995. Species concepts and phylogenetic analysis - introduction. Systematic Botany 20: 555-559.
Davis, J.I. 1996. Phylogenetics, molecular variation, and species concepts. BioScience 46: 502-510.
Davis, J.I. & Nixon, K.C. 1992. Populations, genetic variation, and the delimitation of phylogenetic species. Systematic Biology 41: 421-435.
de Queiroz, K. & Donoghue, M.J. 1988. Phylogenetic systematics and the species problem. Cladistics 4:317-338.
Dobzhansky, T. 1937. What is a species? Scientia: 280-286.
Donoghue, M.J. 1985. A critique of the biological species concept and recommendations for a phylogenetic alternative. Bryologist 88: 172-181.
Ehrlich, P.R. 1961. Has the biological species concept outlived its usefulness? Systematic Zoology 10: 167-176.
Frost, D.R. & Hillis, D.M. 1990. Species in concept and practice: Herpetological applications. Herpetologica 46: 87-104.
Fry, A.J. & Zink, R.M. 1998. Geographic analysis of nucleotide diversity and song sparrow (Aves: Emberizidae) population history. Mole. Ecol. 7: 1303-1313.
Gill, F.B. 1995. Ornithology. New York; W. H. Freeman & Co.
Graybeal, A. 1995. Naming species. Systematic Biology 44: 237-250.
Hazevoet, C.J. 1996. Conservation and species lists: taxonomic neglect promotes the extinction of endemic birds, as exemplified by taxa from eastern Atlantic islands. Bird Conservation International 6: 191-196.
Hennig, W. 1966. Phylogenetic systematics. Chicago; University Illinois Press.
Luckow, M. 1995. Species concepts: assumptions, methods, and applications. Systematic Botany 20: 589-605.
Mallet, J. 1995. A species definition for the Modern Synthesis. Trends Ecology Evolution 10: 294-299.
Mayden, R.L. & Wood, R.M. 1995. Systematics, species concepts, and the evolutionarily significant unit in biodiversity and conservation biology. American Fish Society Symposium 17: 58-113.
Mayr, E. 1942. Systematics and the origin of species. New York; Columbia University Press.
Mayr, E. 1963. Animal species and evolution. Cambridge (Massachusetts); Harvard University Press.
Mayr, E. 1982. The growth of biological thought. Cambridge (Massachusetts); Belknap Press.
Mayr, E. 1993. Fifty years of research on species and speciation. Proceedings California Academy Sciences 48: 131-140.
Miller, A.H. 1941. Speciation in the avian genus Junco. University California Publications in Zoology 44: 173-434.
Moritz, C. 1994. Defining 'evolutionarily significant units' for conservation. Trends Ecology Evolution 9: 373-375.
Nelson, G & Platnick, N.I. 1981. Systematics and biogeography. New York; Columbia University Press.
Paterson, H.E.H. 1985. The recognition concept of species. Transvaal Museum Monographs 4: 21-29.
Nixon, K.C. & Wheeler, Q.D. 1990. An amplification of the phylogenetic species concept. Cladistics 6: 211-223.
Rosen, D.E. 1979. Fishes from the uplands and intermontane basins of Guatemala: Revisionary studies and comparative geography. Bulletin American Museum Natural History 162: 267-376.
Sangster, G., Hazevoet, C.J., van den Berg, A.B. & Roselaar (Kees), C.S. 1997. Dutch avifaunal list: taxonomic changes in 1977-97. Dutch Birding 19: 21-28.
Swarth, H.S. 1920. Revision of the avian genus Passerella with special reference to the distribution and migration of the races in California. University California Publications Zoology 21: 75-224.
Templeton, A.R. 1989. The meaning of species and speciation: a genetic perspective. In: Otte, D. & Endler, J.A. (eds) Speciation and its consequences. Sunderland; Sinauer Associates: 3-27.
Whittemore, A. 1993. Species concepts: a reply to Ernst Mayr. Taxon 42: 573-583.
Yang, S.Y. & Patton, J.L. 1981. Genic variability and differentiation in the Galapagos finches. Auk 98: 230-242.
Zink, R.M. 1994. The geography of mitochondrial DNA variation, population structure, hybridisation, and species limits in the Fox Sparrow (Passerella iliaca). Evolution 48: 96-111.
Zink, R.M. 1996. Bird species diversity. Nature 381: 566.
Zink, R.M. 1997a. Species concepts. Bulletin British Ornithologists Club 117: 97-109.
Zink, R.M. 1997b. Phylogeographic studies of North American birds. In: Mindell, D.P. (ed.) Avian Molecular Evolution and Systematics. New York; Academic Press: 297-320.
Zink, R.M. & McKitrick, M.C. 1995. The debate over species concepts and its implications for ornithology. Auk 112: 701-719.
Fig. 1. Hypothetical phylogeny for five ‘intraspecific’ taxa, and the matrix that indicates their ability to hybridise in sympatry. Reproductive isolation has evolved inconsistently with the history of population differentiation. To see the implications of adopting a BSC, consider the following extension of our example. Imagine that taxa A, B, and C possess a common trait, such as ‘red flowers’ and one is interested in the evolution of this trait. If we ranked A, C, and E as conspecific because they hybridise, one might infer that red flowers evolved independently in A, B, and C, whereas if species names correctly reflected historical relationships, one would learn that red flowers evolved in the common ancestor of A, B, and C.

Fig. 2. (Top) Phylogenetic pattern in the Fox Sparrow Passerella iliaca determined from mtDNA restriction sites (Zink 1994).The taxa schistacea and megarhyncha meet and hybridise in a narrow zone which results in their being placed in the same species (Bottom) under the BSC. However, this makes megarhyncha paraphyletic, which is a useless taxon in modern systematics and evolutionary biology.
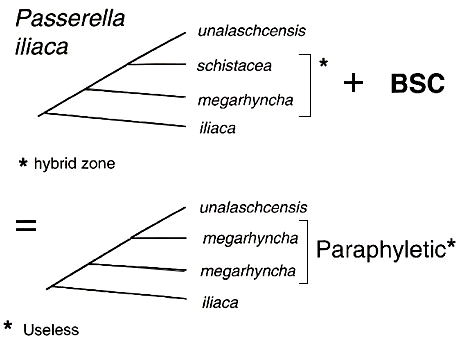
Fig. 3. Consensus cladogram for 20 mitochondrial DNA haplotypes found within the iliaca group of the Fox Sparrow (Zink 1994). Two-letter codes refer to localities shown in Zink (1994) and numbers of individuals with each haplotype shown in parentheses (if more than one). Note that haplotypes from the same locality are often geographically scattered on the tree (e.g., NF [Newfoundland]). Although each haplotype is genetically diagnosable no other evidence exists to support more finely subdividing iliaca into phylogenetic species.
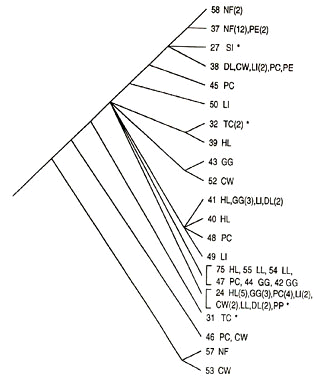
Fig. 4. History of classification of Darwin's finches. After 1900 implementation of the BSC and the resultant 'subspecies concept' reduced the number of recognised biological species, with a concomitant rise in numbers of subspecies. The total number of taxa is less than the sum of species and subspecies because some species consist of only one subspecies. Each point represents a taxonomic revision. Re-classification of these birds with the phylogenetic species concept would likely result in the number of species being equal to or less than the total number of taxa recognised at the present time.