S26.1: Resolution of the debate over species concepts in ornithology: a new comprehensive biologic species concept
Ned K. Johnson1,2, J.V. Remsen Jr.3 & Carla Cicero1
1Museum of Vertebrate Zoology and 2Department of Integrative Biology, University of California, Berkeley, CA 94720-3160, USA, e-mail neddo@socrates.berkeley.edu; 3Museum of Natural Science, Louisiana State University, Baton Rouge, LA 70803, USA
Johnson, N.K., Remsen Jr., J.V. & Cicero, C. 1999. Resolution of the debate over species concepts in ornithology: a new comprehensive biologic species concept. In: Adams, N.J. & Slotow, R.H. (eds) Proc. 22 Int. Ornithol. Congr., Durban: 1470-1482. Johannesburg: BirdLife South Africa.
Proponents of the phylogenetic species concept (PSC) have attacked the biologic species concept (BSC), long a standard in ornithology, on the basis of three principal criticisms: subjective treatment of allopatric populations, improper interpretation of hybridisation, and the recognition of non-historical groups. Each criticism has been overstated and can be countered. First, levels of genetic and vocal divergence in sympatric congeners can be used objectively to determine taxonomic rank of allopatric populations. With respect to hybridisation, many undisputed biologic species of birds long retain the capacity for at least limited interbreeding with other species, even non-sister taxa. Essential, not total, reproductive isolation has been the operational criterion for species status under the BSC. Third, biologic species can reflect historical relationships; incorrect hypotheses of such relationships based on morphologic criteria easily can be replaced by molecular-based estimates. Essential genetic independence resulting from reproductive isolation, the heart of the BSC, is responsible for the evolution of avian biodiversity. The PSC, in contrast, suffers from lethal flaws: the diagnosis of species by phenetic criteria at an unspecified operational level, disregard for the crucial role of isolating mechanisms in the maintenance of genetic discontinuities, and the loss of information on different levels of population interaction that would accompany the elimination of subspecies. Although the overarching validity of the traditional BSC in ornithology is here reaffirmed, an overly-strict interpretation of the BSC by some modern workers suggests that a broadened, more inclusive definition could obviate further debate. To that end we propose a Comprehensive Biologic Species Concept (CBSC) for birds.
INTRODUCTION
Although a number of definitions of species have been proposed in systematic biology during the past half-century, the biologic species concept (BSC) of Dobzhansky (1937) has long held sway for most groups of organisms, including birds (Mayr 1942, 1963). During the past 15 years, however, the theoretical and practical underpinnings of the BSC in ornithology have been strongly criticised, particularly by advocates (Cracraft 1983; McKitrick and Zink 1988; Zink and McKitrick 1995; Zink 1996) of the phylogenetic species concept (PSC). Because the aforementioned papers have elicited minimal response from BSC-oriented ornithologists (Amadon and Short 1992; Snow 1997; Collar 1997), and in view of renewed attacks (Zink 1997) on the BSC as applied to ornithology, we fear that further acquiescence could be misinterpreted as compliance.
We confine our remarks to the applicability of the BSC to birds, a group with obvious and easily studied reproductive isolating mechanisms. We withhold judgment on the appropriateness of the BSC for the multitude of non-avian organisms whose reproductive isolating mechanisms are either poorly understood or unknown (e.g., marine invertebrates) or whose breeding systems are diverse and complex (e.g., plants). That reservation notwithstanding, we believe that the essence of the BSC - both from a philosophical and practical standpoint - is unassailably superior for the treatment of avian species. Therefore, we disagree with Cracraft (1983), McKitrick and Zink (1988), and Zink (1996), who intimate that the supposed popularity of the PSC among workers on other groups of organisms is reason to apply it to birds. Flocks may follow errant shepherds.
We first address the three principal criticisms of the BSC by PSC advocates: subjective treatment of allopatric populations, improper interpretation of hybridisation, and the recognition of non-historical groups. Each criticism has been overstated, can be countered, and none seriously undermines the validity of the BSC. Furthermore, the BSC as portrayed by PSC proponents does not reflect typical treatment of real taxa, a misrepresentation responsible for some of the criticism. Moreover, the increasing availability of molecular-based phylogenies - some of which may conflict with phylogenies based on morphologic evidence - does not require rejection of the BSC. The latter can easily accommodate robust phylogenetic data as they are produced. Second, we review the several severe problems that haunt the PSC when applied to birds. Finally, we argue that perceived problems with the BSC have resulted at least in part from narrowness of interpretation stemming from the sparse wording of its original definition. To eliminate this shortcoming we introduce a more inclusive and strengthened definition that we term the Comprehensive Biologic Species Concept (CBSC).
RESPONSE TO CRITICISMS OF THE BIOLOGIC SPECIES CONCEPT
Allopatric populations can be assessed objectively
As recognised long ago by Mayr (1942), taxonomic ranking of allopatric populations has long posed a ‘problem’ for the BSC for the obvious reason that no ‘test of sympatry’ is possible. The crux of the matter is that allopatric populations exhibit varying levels of divergence from their closest relatives, ranging from those weakly distinguishable by any criteria, through intermediate populations that defy allocation to either the subspecies or species category, to populations so different that their ranking as species is beyond debate. PSC proponents have argued that treatment of allopatric populations under the BSC must therefore be subjective and have used this as a major reason to discount the BSC as it applies to birds.
Technology developed over the last half century, however, including spectrographic analysis of tape recordings of voice, videotapes of displays, and assessment of genotypic differences through allozyme electrophoresis and various DNA analyses, offers objective, quantitative, explicit and repeatable means by which taxonomic rank of allopatric populations can be assigned. For example, through a quantitative comparison of both acoustic and visual components of courtship displays in allopatric populations of the Sage Grouse Centrocercus urophasianus, Young et al. (1994) discovered a complex of unique features confined to populations occupying an isolated montane basin near Gunnison, Colorado. Certain of these special secondary sexual characteristics are correlated with mating success in other populations. In addition to differences in behaviour, the isolated populations also possessed distinctive feather morphology and diverged strongly in body size from other allopatric populations of Sage Grouse. These findings prompted Young et al. (1994) to choose rapid speciation through intense sexual selection as the evolutionary engine driving the system. The American Ornithologists’ Union (1998) concluded that the Gunnison populations represent a distinct (still unnamed) species. This study vividly exemplifies how modern, sophisticated data can enable objective judgments on the systematic status of allopatric populations. The comparative, quantitative analysis of songs and calls, especially in taxa in which vocalizations are innate, also has great potential for clarifying species limits in allopatric populations (Isler et al. 1998). Finally, genetic analysis of allopatric populations allows for the quantitative and explicit determination of levels of divergence, providing corroborative evidence of species status ( Zink et al. 1988, and Zink and Dittmann 1991 [for Pipilo]; Peterson 1992 [for Aphelocoma]; Johnson 1995 [for Vireo]; Cicero 1996 [for Baeolophus]).
Hybridisation does not automatically confer conspecificity under the BSC
Proponents of the PSC conclude that hybridisation provides direct evidence for the breakdown of reproductive isolating mechanisms and loss of genetic integrity, requiring that hybridising ‘species’ be lumped under the BSC. In practice, however, this has not been the case. Many avian taxa, even non-sisters, are known to interbreed to varying degrees, yet they are not ranked as conspecifics under the BSC. Although the essence of any biologic species is its genetic independence, thus allowing evolution on a trajectory distinct from all other existing species, such genetic independence need not be complete, and many avian species separated for millennia retain the ability to interbreed (Prager and Wilson 1975). Grant and Grant (1992) stated that ‘over 10% of recognised avian species are estimated to have engaged in hybridisation producing viable offspring.’ Evidently, the comparatively sluggish development of postmating isolating mechanisms in birds has increased their opportunity for introgressive hybridisation (Grant and Grant 1997). Nonetheless, It is important to recognise the distinction between retention of the ability to hybridise and demonstrated hybridisation. Many avian taxa that are well-known to hybridise under the artificial conditions of captivity do so rarely or never in the wild.
Thus, contrary to the interpretation of the BSC by proponents of the PSC, genetic exchange between essentially reproductively isolated forms of birds treated as species has long been recognised and accepted by avian systematists. Examples of such hybridisation range from that recorded occasionally between undoubted close relatives (Gambel’s Quail Callipepla gambeli X California Quail C. californica, Henshaw 1885) to the rare mating and successful production of offspring in the wild by indisputable non-congeners such as the Common Goldeneye Bucephala clangula X Hooded Merganser Lophodytes cucullatus (Kortright 1943; Mayr and Short 1970). Moreover, even extensive hybridisation has not led the AOU (1983, 1998) to lump the following pairs of taxa: American Black Duck A. rubripes X Mallard A. platyrhynchos; Western Gull Larus occidentalis X Glaucous-winged Gull Larus glaucescens; Blue-winged Warbler Vermivora pinus X Golden-winged Warbler V. chrysoptera; Townsend’s Warbler Dendroica townsendi X Hermit Warbler Dendroica occidentalis; Collared Towhee Pipilo ocai X Spotted Towhee Pipilo maculatus; Rose-breasted Grosbeak Pheucticus ludovicianus X Black-headed Grosbeak P. melanocephalus; and Lazuli Bunting Passerina amoena X Indigo Bunting P. cyanea. In general, hybridisation per se has not been the ironclad criterion for conspecificity under the BSC that PSC proponents would have one believe. Whether interbreeding is free (= nonassortative mating) or mating is assortative to varying degrees between interacting populations will importantly influence taxonomic treatment under the BSC. What remains to be decided by either BSC or PSC proponents is the degree of hybridisation allowed before two groups of interacting populations are deemed conspecific.
Two complexes of taxa, however, deserve special comment. The ‘Yellow-shafted’ and ‘Red-shafted’ flickers Colaptes auratus auratus and C. a. cafer hybridise broadly in the interrupted woodland across the North American Great Plains, and for this reason are treated by the AOU (1983, 1998) as a single species under the BSC. Using mtDNA assays, however, Moore et al. (1991) found that these taxa probably are not sisters. We agree with Zink (1996a) that their lumping (AOU 1983) may incorrectly reflect history. Nonetheless, because only some of the taxa in the complex have been analyzed thus far by molecular systematic techniques, the AOU (1998) is justified in maintaining them provisionally as one species. When the mtDNA of all of the various geographic forms have been analyzed, we anticipate a classification that properly recovers phylogeny without abandoning the BSC.
The ‘Myrtle’ and ‘Audubon’s’ warblers Dendroica coronata coronata and D. c. auduboni, respectively illustrate a similar situation. Because they apparently hybridise freely at several sites over a long, narrow zone in the Canadian Rockies (Hubbard 1969), the AOU (1983) treated them as a single species under the BSC. As in the flickers, this example also may represent a non-historical alignment of species taxa (Zink and McKitrick 1995). However, splitting them would be premature in the absence of a modern systematic analysis of all relevant taxa. Thus, nomenclatural changes cannot be justified at this time. Neither in the flickers nor the warblers is it necessary to abandon the BSC to reduce emphasis on hybridisation and to recover history, once all taxa have been surveyed by molecular approaches. Thus, essential genetically cohesive entities can continue to be recognised in each group under the BSC.
PSC proponents operating under cladistic philosophy give little weight to degree of interbreeding as a species criterion because they view interbreeding as the ‘ancestral state’ of the character ‘ability to interbreed.’ Retention of the ability to interbreed is thus seen as ‘primitive,’ and lack of the capacity to interbreed is the ‘derived character state.’ Individuals interbreeding in populations that have always been in contact because they evolved together from a common ancestor are most certainly demonstrating the primitive character state in a cladistic sense. As speciation proceeds, however, repeated geographic shuffling of clusters of individuals in subpopulations that are first totally isolated, then recombined with sister populations, then again geographically isolated, causes the lineages to reticulate so that the designation of ‘primitive’ versus ‘derived’ becomes meaningless and impossible to apply. Certainly, interbreeding upon secondary contact after a substantial period of geographic isolation logically could not be called ‘ancestral.’ Instead, reticulation would produce a series of reversals in the character ‘ability to interbreed,’ whose alternating state would be of dubious value in reconstructing phylogenetic history. Moreover, a serious technical problem attends the treatment of ‘the ability to interbreed’ as a ‘character’ in the usual sense because it cannot be measured independently among individuals.
Whether or not the ability to interbreed can be regarded properly as a formal character, we underscore the fact that degree of interbreeding continues to be an excellent predictor of relatedness, despite the existence of a relatively few intractable cases. Indeed, it is difficult to conceive of a more effective general indicator of close relationship, given the multitude of continuing problems with character definition and analysis. More importantly, we regard as absurd the outlawing of interbreeding, the most important process in gene flow and population differentiation, because it is defined as ‘primitive’ in some narrow, cladistic sense. Although the role of hybridisation in defining biologic species may have been overemphasised, the fundamental concept of essentially independent genetic entities embodied by the BSC can remain intact while the ability to interbreed is accorded lesser importance, the status it has had in actual practice.
Information on phylogenetic history can be incorporated into existing classifications without abandoning the BSC
Although Hennig (1950, 1966) was the first to articulate a rigorous set of rules and assumptions by which common ancestry can be elucidated, he was by no means the first to show interest in phylogeny. Data from traditional studies of morphology, behaviour, and distribution of birds, combined with results from allozyme and DNA techniques, have provided a wealth of information from which to deduce phylogenetic history, with a surprising degree of concordance with results from Hennigian phylogenetic systematics. Many studies in which the PSC was not applied have revealed information about relationships and, hence, common ancestry and phyletic history. We did not need the PSC to inform us, for example, that the Nashville Warbler Vermivora gutturalis and Virginia’s Warbler Vermivora virginiae are closest living relatives in the Hennigian sense or that the Island Scrub Jay Aphelocoma insularis evolved from mainland ancestors.
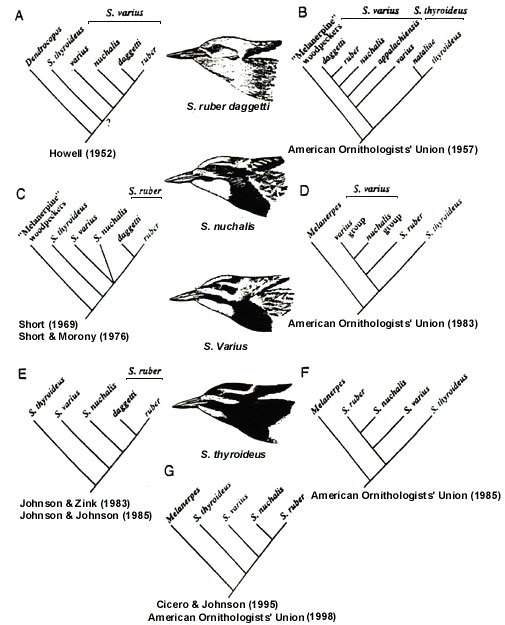
Many active laboratories around the world are currently providing a wealth of new data on the molecular systematics of birds. As these data are obtained they can be applied to continuing problems in avian evolutionary history and relationships of taxa. Most importantly, the new information can be used either to corroborate or to refute existing classifications without adopting the PSC. A clear example is provided by the changing systematic treatment of sapsuckers Sphyrapicus; (Fig. 1). Current classification (AOU 1998) recognises four biologic species: S. thyroideus, including S. t. thyroideus and S. t. nataliae; S. varius, including S. v. varius and S. v. appalachiensis; S. nuchalis (monotypic); and S. ruber, including S. r. ruber and S. r. daggetti. Howell (1952) provided the first detailed study of this group, in which he merged ruber and nuchalis into varius because of overall similarities in plumage, natural history, and essentially allopatric distributions. Short (1969) and Short and Morony (1970), however, noted the lack of free interbreeding in sympatry and used this as evidence for species status within the superspecies S. varius. Short and Morony (1970) also posited that the three taxa are ‘barely specifically distinct,’ suggesting an unresolved trichotomy (Johnson and Zink 1983). The American Ornithologists' Union (1983) favoured S. ruber as a distinct species but maintained nuchalis as a subspecies of S. varius. Using allozymes, Johnson and Zink (1983) showed that ruber and nuchalis are essentially identical genetically despite their phenotypic dissimilarity, whereas varius, which closely resembles nuchalis in plumage pattern, is distinct allozymically. Although the genetic data alone might suggest that ruber and nuchalis are conspecific, Johnson and Johnson (1985) demonstrated that these taxa show a strong tendency toward positive assortative mating where they occur in local sympatry, principally in south-central Oregon and northeastern California. Thus, subsequent treatment (AOU 1985) split nuchalis and varius but retained S. ruber as a species. Using mitochondrial DNA sequences, Cicero and Johnson (1995) corroborated the results from allozymes and supported the species-level treatment of the three taxa. The species status of S. thyroideus has never been questioned, although its sequential position has been debated. Both the allozyme and mtDNA studies firmly establish the ancestral position of S. thyroideus and the relatively derived positions of S. varius, S. nuchalis, and S. ruber within the genus.
The current classification allows species status for two interbreeding forms (S. ruber and S. nuchalis) despite their hybridisation because (1) there is no sign that the hybridisation is influencing their respective evolutionary trajectories, and (2) significantly large populations in sympatry are maintaining their essential cohesiveness through predominantly assortative mating. Again, hybridisation per se has not required conspecificity for these taxa. Furthermore, the incorporation of genetic data allows them to be regarded as sister taxa despite the fact that S. nuchalis more closely resembles the non-sister taxon, S. varius, than it does its closest relative, S. ruber. Although S. thyroideus, S. varius and S. ruber each contain two weakly differentiated subspecies, because they are diagnosible units, PSC-proponents would elevate these to species. Nothing would be gained by bestowing species status on such poorly-divergent forms, and no historical information is lost by recognising four species under the BSC with the following relationship: (thyroideus (varius (nuchalis, ruber))). Finally, it is important to note that degree of interbreeding correctly predicts phylogeny in this example.
CRITICISMS OF THE PHYLOGENETIC SPECIES CONCEPT
Phylogenetic species are not inherently phylogenetic
The expression ‘phylogenetic species’ is a misnomer. The application of the concept as envisioned by Cracraft (1989) involves nothing more than drawing boundaries around clusters of individuals with shared characters that are assumed, not proved, to have a genetic basis. This exercise reveals nothing about phylogeny per se and, therefore, could be applied equally well to inanimate objects (see below). In contrast, the BSC can be applied only to living organisms. Even Cracraft (1996) stated explicitly that the PSC is not based on phylogeny and thus is inappropriate in classifications with a phylogenetic basis.
Phylogenetic species are subjectively defined, physical entities
Vocalisations, colouration, and displays serve in mate attraction and therefore maintain genetic cohesion of avian populations. These features are easily studied in free-living individuals, enabling the recognition of mated pairs. Consequently, the birds themselves reveal the particular individuals with which they prefer to mate. Importantly, the same behaviours and signals that unite members of interbreeding groups also simultaneously reduce or prevent pairing with members of other cohesive populations. In this fashion, biologic species are based on objective biologic criteria. In contrast to such biologic species, Cracraft’s (1989) phylogenetic species are physical entities (‘diagnosable basal units’), defined by either phenetic or cladistic criteria. Therefore, when strictly interpreted, they lack properties that clearly distinguish them from groups of inanimate objects, as in the following example. A prisoner in a quarry breaking rocks with a sledgehammer can easily divide a lifeless boulder (parent material) into daughter (= sister) ‘species’, each possessing diagnosable phenetic features. Another hammer blow on one rock could produce another pair of diagnosable sister species. Back in his cell, if he were cladistically inclined, he might draw a cladogram on the wall above his cot which could precisely show the history of the lineage represented by the original boulder and the sequence of splitting events though time that produced the three descendant ‘phylogenetic species.’ This facetious example serves to remind that the fundamental biological nature of entities recognised by the BSC is one of its outstanding features.
Although the PSC is not inherently cladistic (Cracraft 1996), many advocates of this concept adhere to cladistic philosophy. The diagnosis of phylogenetic species by cladistic rather than phenetic criteria does not necessarily produce defined units with an historical or phylogenetic basis, however, because of problems with the practical application of cladistic methodology. For example, much evidence from birds indicates that single lineages at early stages of divergence may typically be of mixed ancestry. In these examples of reticulation, tidy dichotomous branches, the foodstuff of cladistics, could easily incorrectly reflect evolutionary history. Instead, a changing network of intertwining individuals would more accurately depict such diverging populations during early speciation. Moreover, excessive worship of Hennig’s catechism binds researchers to a set of procedures that either may be misleading at best or non-biological at worst, and infatuation with every detail of a particular philosophy easily restricts or excludes novel conceptual thinking about how evolution might occur. Finally, all too often researchers blindly accept the validity of published phylogenetic trees. As even a cursory inspection of the recent literature will show, such trees are nothing more than hypotheses of varying rigor whose topologies are typically beset with non-trivial problems of their own (Hillis et al. 1996). Regardless of the power of existing algorithms and the ability of techniques such as bootstrapping to test confidence limits of branch lengths and nodes, their reliability to extract accurately phylogenetic signal from data sets remains conjectural (Sanderson 1995). Healthy skepticism of the robustness of such trees continues to be the only realistic stance (Helm-Bychowski and Cracraft 1993).
The operational level for the ‘diagnosability’ of phylogenetic species is ambiguous
Whereas the BSC focuses on fundamental biological processes, namely amount of gene flow and degree of genetic independence, the PSC focuses on pattern. A fundamental weakness of such a pattern-based definition is arbitrariness. At what level of diagnosability should the phylogenetic species be defined? And, if a level can be chosen, is it biologically defensible? The heart of the problem is that relative diagnosability exists as a continuum, and because the cutoff level for the definition of phylogenetic species cannot be specified on biologic grounds the PSC is rendered non-operational. In the extreme case, because every single organism is phenetically and genetically unique, the ‘diagnosable basal unit’ becomes either small clusters of individuals or the individual itself and the PSC is thereby reduced to absurdity.
By eliminating subspecies, the PSC discards valuable information on levels of evolutionary interaction
Application of the PSC would elevate all diagnosable BSC subspecies (estimated by Zink [1996b] to be approximately 50% of current BSC subspecies) to phylogenetic species. Although apparently innocuous at first glance, this action would have several consequences. Mayr (1993), for example, was concerned with the unwieldiness of the increased absolute number of taxa, and Snow (1997) predicted inevitable taxonomic instability. Neither of these reservations, however, would justify rejection of the PSC if it indeed resulted in a species list of greater biologic reality than that found under the BSC. Instead, we recognise an additional, heretofore unmentioned problem that results from the fact that allopatric populations exist at varying levels of genetic and morphologic divergence and in differing patterns of spatial arrangement. Some clusters of populations with clearly marked characters, currently called subspecies under the BSC, are probably species in the making and thus are basal evolutionary units in the sense of Cracraft (1989). Their elevation to species under the PSC would present no problem if they were insular or otherwise disjunctly allopatric, i. e., without integradation with neighbouring subspecies. BSC subspecies with zones of integradation with adjacent subspecies, however, present a serious issue that the PSC fails to consider. If such BSC subspecies are elevated to phylogenetic species status, the intergradient populations between them, which by definition belong to neither subspecies and are therefore not basally diagnosable, are left in taxonomic limbo. Furthermore, when populations representing BSC subspecies with primary intergradation are elevated to phylogenetic species they now have the same taxonomic status as species engaging in rare or at least infrequent secondary contact and hybridisation. The lumping under the PSC of these two distinct biological phenomena, primary intergradation and secondary contact with hybridisation, would result in a major loss of information on kinds of evolutionary interactions of populations. In addition, assessed levels of ‘interspecific’ hybridisation also would be artificially inflated.
In practice, PSC advocates elevate to species only obvious, well-marked BSC subspecies or subspecies groups without diagnosing all populations
The treatment of geographic differentiation of forms in the Brown Towhee complex (Pipilo) is a pertinent example of the arbitrary fashion in which geographic variation is treated under the PSC. Using allozyme and morphometric data, Zink (1988) concluded that coastal populations (crissalis group of the ‘Brown Towhee’) were more closely allied to the Abert’s Towhee P. aberti than to southwestern interior populations (fuscus group of the ‘Brown Towhee’), with which they had long been combined by the AOU (up to and including AOU 1983). Zink and Dittmann (1991) and Zink et al. (1998) corroborated the allozyme results with a survey of mitochondrial DNA restriction fragments and mtDNA base sequences, respectively. Their findings are significant not only for their better estimates of the probable evolutionary history of the complex, but also because they reveal the ease with which the taxonomic status of allopatric populations can be clarified by molecular systematic approaches without resorting to the PSC. Cracraft (1989), however, assumed that Zink’s (1988) corrected phylogeny required abandonment of the BSC and improperly used the example as a cause celebre for the PSC. More seriously, Cracraft (1989) relegated to the dustbin the eight subspecies of the crissalis group and the 11 subspecies of the fuscus group, all carefully analysed by Davis (1951), because ‘None of these subspecies is apparently diagnosably distinct.’ The latter conclusion was offered in the absence of any analysis of these subspecies to test their diagnosability. (Such is common practice of PSC advocates—apply it to the obvious, borderline biological species but not to apply it thoroughly to all taxa involved.) Furthermore, Cracraft (1989) overlooked the fact that Zink (1988) clearly showed that at least one sample from Baja California (subspecies unnamed) was diagnosably distinct from other examples of the crissalis group from farther north in California. Cracraft’s cavalier dismissal of nearly two-score taxa is indefensible because the burden of proof of non-diagnosability rests on the shoulders of those who would reject these taxa. This is not to state that modern analytical techniques would validate all these subspecies, but only to note that at the very least they deserve a fair trial.
Zink’s (1997) discussion of the Fox Sparrow Passerella iliaca, currently treated as one species under the BSC, again exposes the fact that the PSC only considers variation at the level of species or near-species according to the BSC. Zink noted that populations of this sparrow fall into four basic groups that he would call phylogenetic species. Although we regard the data as inconclusive as to whether four species should be recognised (given their essential allopatry and inadequately studied patterns of haplotype distribution, hybridisation and vocal variation), we are concerned here with another issue. Zink (1997) noted that the four basic groups have been ‘partitioned by taxonomists into 18 subspecies.’ Surprisingly, he disregarded this extensive variation in morphology and colour when applying the PSC to this complex. Even with refined current techniques, at least some of these subspecies are diagnosable. What is to become of them if they are considered, wholly subjectively, not to represent basal evolutionary units? Therefore, this treatment under the PSC focused only on the obvious near-species, as far as diagnosable taxa are concerned, and ignored what might be true basal evolutionary units below that level.
Thus, significant variation has not been accommodated in several prominent applications of the PSC, even though it could have been, resulting in a major loss of information on geographic variation. In contrast, a major attribute of the BSC is its appreciation of intraspecific geographic variability and the recognition of complex patterns of change in size, colouration, voice and genetic population structure at different taxonomic and populational levels. Especially informative are levels of population differentiation analysed with reference to the natural geographic setting of climatic barriers and habitat discontinuities, and associated evolutionary processes, that gave rise to the patterns in the first place.
We conclude that the serious weaknesses of the phylogenetic species concept render it unsuitable for ornithology. Because it is based on the arbitrary criterion of phenetic diagnosability, it is non-biological and retrogressive. The PSC offers no advance over the traditional biologic species concept and is therefore unnecessary.
A COMPREHENSIVE BIOLOGIC SPECIES CONCEPT FOR BIRDS
Although we have concentrated our remarks on the BSC and PSC, several other species concepts have been prominent in the literature of this century. These concepts are variously redundant, are not mutually exclusive, and differ more in viewpoint than substance in their emphasis on particular aspects of species. Moreover, definitions of at least certain of these concepts are various mixtures of process (how species are formed) and eventual pattern (the resulting species ‘entity’). We submit that the important attributes of all these concepts can be incorporated into a modern, comprehensive Biologic Species Concept, as is evident from comments in the list below. Although we repeat Avise’s (1994) list of definitions, the remarks following each concept are ours.
(1) Biologic Species Concept (BSC) of Dobzhansky (1937): ‘Species are systems of populations: the gene exchange between these systems is limited or prevented by a reproductive isolating mechanism or perhaps by a combination of several such mechanisms.’ In our view this concept holds primacy. Note that it stresses the critical role of reproductive isolating mechanisms in maintaining the essential genetic integrity of species but that it also allows for limited gene exchange among species. The inclusion of the expression ‘gene exchange’ is also important because it offers an albeit cryptic reference to reproductive generations and, hence, phylogeny, because genes can only be exchanged between parents and offspring. Thus, although the word ‘phylogeny’ is not an explicit part of the definition of the BSC, the concept has an undeniably phylogenetic component that heretofore has gone unrecognised and, therefore, unappreciated.
(2) Evolutionary Species Concept (ESC) of Simpson (1951) refers to ‘a lineage (ancestral-descendant sequence of populations) evolving separately from others and with its own unitary evolutionary role and tendencies.’ This paleontological viewpoint emphasises distinct evolutionary trajectories of lineages through time. Although the mechanisms by which species evolve separately and maintain their ‘unitary evolutionary role’ are not specifically mentioned, the definition does not exclude such mechanisms and implicitly recognises their occurrence. The gist of this concept easily can be absorbed into the BSC.
(3) Recognition Species Concept (RSC) of Paterson (1985, 1993) refers to ‘that most inclusive population of individual biparental organisms which share a common fertilisation system.’ This concept underscores the importance of fertilisation systems, including, especially, mate recognition, as a positive force assuring the assortment of individuals that circumscribe species. Rather than emphasise isolation of different species by particular mechanisms, the RSC values the means by which individuals keep species cohesive through time. We regard the principal thrust of the RSC as a viewpoint rather than a distinct philosophy, a stance that is subsumed without difficulty under the BSC.
(4) Cohesion Species Concept (CSC) of Templeton (1989) is defined as ‘the most inclusive population of individuals having the potential for cohesion through intrinsic cohesion mechanisms.’ As Avise (1994) noted, ‘The major classes of cohesion mechanisms are genetic exchangeability . . . And demographic exchangeability . . .’ This concept emphasises the importance of genetic processes in gluing together individuals of species. Again, genetic cohesion is implicit in the traditional BSC, and we find this concept to be unnecessary.
(5) Phylogenetic Species Concept (PSC) of Cracraft (1983) was defined as ‘a monophyletic group composed of the ‘smallest diagnosable cluster of individual organisms within which there is a parental pattern of ancestry and descent.’ The PSC has been discussed at length above, where we expose its serious problems. In rejecting the PSC, we remind that species defined under the BSC have the capacity to incorporate phylogenetic information whether or not such is explicitly mentioned in the definition.
(6) Concordance Principles Concept (CPC) of Avise and Ball (1990) is ‘a suggested means of recognising species by the evidence of concordant phylogenetic partitions at multiple independent genetic attributes’ (Avise 1994). While accepting the basic premise of the BSC (Avise 1994), this concept invokes the importance of intrinsic reproductive barriers in the evolution of genealogical concordance within species. Although it exposes the fact that reticulation must be overcome before essential genetic integrity (speciation) can develop, we reason that the evolution of such concordance would be expected during the formation of biologic species whether or not explicitly stated in traditional BSC definitions.
We suggest that disagreements over species concepts in birds stem principally from two sources. First, an overly narrow interpretation of the BSC has led to the false assumptions that biologic species cannot engage in interspecific hybridisation without being combined, and that the BSC leads to an incorrect interpretation of phylogenetic history because of hybridisation of non-sister taxa. Although hybridisation has traditionally been assumed to occur between sisters, such hypotheses can be tested now with molecular data, as stated above. In many instances, genetic data have verified that the hybridising taxa are indeed sisters and that free interbreeding is a fairly reliable indicator of sister status. Second, narrow definitions of species are incompatible with the inherent complexity of avian speciation because such definitions can never accommodate all possible interactions of natural populations. We therefore conclude that the only definition of species that approaches evolutionary reality must be much more generalised and inclusive than any offered heretofore. Such a broadened, modernised species concept is eminently applicable to birds because it can incorporate the essences of the ESC, RSC, CSC, PSC, and CPC without abandoning the overarching validity of the existing BSC. Thus, we propose for birds the following Comprehensive Biologic Species Concept (CBSC): An avian species is a system of populations representing an essentially monophyletic, genetically cohesive, and genealogically concordant lineage of individuals that share a common fertilisation system through time and space, represent an independent evolutionary trajectory, and demonstrate essential but not necessarily complete reproductive isolation from other such systems.
REFERENCES
Amadon, D. & Short, L.L. 1992. Taxonomy of lower categories-suggested guidelines. Bulletin British Ornithologists’ Club Centenary Supplement 112A: 11-38.
American Ornithologists’ Union. 1957. Check-list of North American birds. 5th edition. Baltimore, Maryland; The Lord Baltimore Press, Inc.: 691pp.
American Ornithologists’ Union. 1983. Check-list of North American birds. 6th edition. Lawrence, Kansas; Allen Press, Inc.: 877pp.
American Ornithologists’ Union. 1985. Thirty-fifth supplement to the American Ornithologists’ Union Check-list of North American birds. Auk 102: 680-686.
American Ornithologists’ Union. 1995. Fortieth supplement to the American Ornithologists’ Union Check-list of North American birds. Auk 112: 819-830.
American Ornithologists’ Union. 1998. Check-list of North American birds. 7th edition. American Ornithologists’ Union, Washington, D. C.: 829pp.
Avise, J.C. 1994. Molecular markers, natural history and evolution. New York, New York; Chapman & Hall: 511pp.
Avise, J.C. & Ball, R.M., Jr. 1990. Principles of genealogical concordance in species concepts and biological taxonomy. Oxford Surveys in Evolutionary Biology 7: 45-67.
Cicero, C. 1996. Sibling species of titmice in the Parus inornatus complex. University of California Publications in Zoology 128: 1-217.
Cicero, C. & Johnson, N.K. 1995. Speciation in sapsuckers (Sphyrapicus): III. Mitochondrial-DNA sequence divergence at the cytochrome-B locus. Auk 112: 547-563.
Collar, N.J. 1997. Taxonomy and conservation: chicken and egg. Bulletin British Ornithologists’ Club 117: 122-136
Cracraft, J. 1983. Species concepts and speciation analysis. Current Ornithology 1: 159-187.
Cracraft, J. 1989. Speciation and its ontology: the empirical consequences of alternative species concepts for understanding patterns and processes of differentiation. In: Otte, D. & Endler, J.A. (eds.) Speciation and its consequences. Sunderland, Massachusetts; Sinauer Associates, Inc.: 28-59.
Cracraft, J. 1996. (Review of) The taxonomy and species of birds of Australia and its Territories, by L. Christidis and W. E. Boles. Auk 113: 973-974.
Davis, J. 1951. Distribution and variation of the brown towhees. University of California Publications in Zoology 52: 1-119.
Dobzhansky, T. 1937. Genetics and the origin of species. New York, New York; Columbia University Press: 364pp.
Grant, P.R. & Grant, B.R. 1992. Hybridisation of bird species. Science 256: 193-197.
Grant, P.R. & Grant, B.R. 1997. Genetics and the origin of bird species. Proceedings of the National Academy of Sciences, USA 94: 7768-7775.
Helm-Bychowski, K. & Cracraft, J. 1993. Recovering phylogenetic signal from DNA sequences: relationships within the corvine assemblage (Class Aves) as inferred from complete sequences of the mitochondrial DNA cytochrome-b gene. Molecular Biology & Evolution 10: 1196-1214.
Hennig, W. 1950. Grundzuge einer Theorie der phylogenetischen Systematik. Berlin; Deutscher Zentralverlag: 370pp.
Hennig, W. 1966. Phylogenetic systematics. Urbana, Univ. Illinois Press: 263pp.
Hillis, D.M., Mable, B.K., Larson, A., Davis, S.K. & Zimmer, E.A. 1996. Nucleic acids IV: sequencing and cloning. In: Hillis, D.M., Moritz, C. & Mable, B.K. (eds) Molecular systematics, second edition; Sunderland, Massachusetts; Sinauer Associates, Inc.: 321-381.
Howell, T.R. 1952. Natural history and differentiation in the Yellow-bellied Sapsucker. Condor 54: 237-282.
Hubbard, J.P. 1969. The relationships and evolution of the Dendroica coronata complex. Auk 86:393-432.
Isler, M.L., Isler, P.R. & Whitney, B.M. 1998. A methodology for the use of vocalizations in establishing species limits in antbirds (Passeriformes: Thamnophilidae). Auk 115: 577-590.
Johnson, N.K. 1995. Speciation in vireos. I. Macrogeographic patterns of allozymic variation in the Vireo solitarius complex in the contiguous United States. Condor 97: 903-919.
Johnson, N.K. & Johnson, C.B. 1985. Speciation in sapsuckers (Sphyrapicus): II. Sympatry, hybridisation, and mate preference in S. ruber daggetti and S. nuchalis. Auk 102: 1-15.
Johnson, N.K. & Zink, R.M. 1983. Speciation in sapsuckers (Sphyrapicus): I. Genetic differentiation. Auk 100: 871-884.
Mayr, E. 1942. Systematics and the origin of species. New York, New York; Columbia University Press: 334pp.
Mayr, E. 1963. Animal species and evolution. Cambridge, Massachusetts; The Belknap Press of Harvard University Press: 797pp.
Mayr, E. 1993. Fifty years of research on species and speciation. Proceedings of the California Academy of Sciences 48: 131-140.
McKitrick, M.C. & Zink, R.M. 1988. Species concepts in ornithology. Condor 90: 1-14.
Moore, W.S., Graham, J.H. & Price, J.T. 1991. Mitochondrial DNA variation in the Northern Flicker (Colaptes auratus). Molecular Biology and Evolution 8: 327-344.
Paterson, H.E.H. 1985. The recognition concept of species. In: Vrba, E.S. (ed.) Species and speciation. Pretoria, South Africa; Transvaal Museum Monograph No. 4: 21-29.
Paterson, H.E.H. 1993. Evolution and the recognition concept of species, collected writings. In McEvey, S.F. (ed.) Baltimore, Maryland; John Hopkins University Press: 234pp.
Peterson, A.T. 1992. Phylogeny and rates of molecular evolution in the Aphelocoma jays. Auk 109:133-147.
Prager, E.M. & Wilson, A.C. 1975. Slow evolutionary loss of the potential for interspecific hybridisation in birds: a manifestation of slow regulatory evolution. Proceedings of the National Academy of Sciences, U.S.A. 72: 200-204.
Sanderson, M.J. 1995. Objections to bootstrapping phylogenies: a critique. Systematic Biology 44: 299-320.
Short, L.L. 1969. Taxonomic aspects of avian hybridisation. Auk 86: 84-105.
Short, L.L. & Morony, J.J. 1970. A second hybrid Williamson’s X Red-naped Sapsucker and an evolutionary history of sapsuckers. Condor 72: 310-315.
Simpson, G.G. 1951. The species concept. Evolution 5: 285-298.
Snow, D.W. 1997. Should the biological be superceded by the phylogenetic species concept? Bulletin British Ornithologists’ Club 117: 110-121.
Templeton, A.R. 1989. The meaning of species and speciation: a genetic perspective. In: Otte, D. & Endler, J.A. (eds) Speciation and its consequences. Sunderland, Massachusetts; Sinauer Associates, Inc.: 3-27.
Young, J.R., Hupp, J.W., Bradbury, J.W. & Braun, C.E. 1994. Phenotypic divergence of secondary sexual traits among sage grouse, Centrocercus urophasianus, populations. Animal Behaviour 47: 1353-1362.
Zink, R.M. 1988. Evolution of brown towhees: allozymes, morphometrics and species limits. Condor 90: 72-82.
Zink, R.M. 1996a. Species concepts, speciation, and sexual selection. Journal of Avian Biology 27: 1-6.
Zink, R.M. 1996b. Bird species diversity. Nature 381: 566.
Zink, R.M. 1997. Species concepts. Bulletin British Ornithologists’ Club 117: 97-109.
Zink, R.M. & Dittmann, D.L. 1991. Evolution of brown towhees: mitochondrial DNA evidence. Condor 93: 98-105.
Zink, R.M. & McKitrick, M.C. 1995. The debate about species concepts and its implications for ornithology. Auk 112: 701-719.
Zink, R.M., Weller, S.J. & Blackwell, R.C. 1998. Molecular phylogenetics of the avian genus Pipilo and a biogeographic argument for taxonomic uncertainty. Molecular Phylogenetics and Evolution 10: 191-201.
Fig. 1. Cladograms illustrating different treatments of sapsuckers (Sphyrapicus) over the past one-half century. Only taxa considered by each author are included. The sequence of taxa, from ancestral to derived, is based on our best interpretation of the data. Drawings show anterior plumage patterns of males.
